ExSTraCS stands for “Extended Supervised Tracking and Classifying System”. ExSTraCS is our own Michigan-Style LCS algorithm geared explicitly towards supervised learning-based data mining, classification, prediction, and knowledge discovery, particularly in the context of bioinformatics, and epidemiological problem domains. Click on the ExSTraCS logo to download the latest version of the free, open source software. ExSTraCS is coded in Python 2.7, it’s cross-platform, and it is run from the command line.
Below we have included the following:
1.) Links to the research publications that introduce, compare, and evaluate ExSTraCS.
2.) Links to complete users guides for both ExSTraCS 1.0 and 2.0.
3.) A schematic of ExSTraCS 2.0, followed by a step-wise summary of the algorithm.
ExSTraCS was developed at Dartmouth College in the lab of Jason H. Moore funded by NIH grants AI59694, LM009012, LM010098, EY022300, LM011360, CA134286, and GM103534. Development and support for ExSTraCS continues at the University of Pennsylvania.
Relevant Publications
Continuous endpoint data mining with ExSTraCS: a supervised learning classifier system.
Ryan Urbanowicz, Nirajan Ramanand, and Jason Moore
Proceedings of the 17th annual conference companion on Genetic and evolutionary computation. ACM Press. 2015.
Retooling fitness for noisy problems in a supervised Michigan-style learning classifier system.
Ryan Urbanowicz, and Jason Moore
Proceedings of the 17th annual conference companion on Genetic and evolutionary computation. ACM Press. 2015.
ExSTraCS 2.0: Addressing Scalability with a Rule Specificity Limit in a Michigan-Style Supervised Learning Classifier System for Classification, Prediction, and Knowledge Discovery.
Ryan Urbanowicz and Jason Moore
Evolutionary Intelligence. 8.2-3, 89-116. 2015
Ryan Urbanowicz
SIGEVOlution Newsletter 7(2-3). pp. 3-11. 2014
ExSTraCS 1.0: An Extended Michigan-Style Learning Classifier System for Flexible Supervised Learning, Classification, and Data Mining.
Ryan Urbanowicz, Gediminas Bertasius, and Jason Moore
Parallel Problem Solving in Nature (PPSN XIII). Springer International Publishing. pp. 221-221. 2014.
Users Guides
ExSTraCS Schematic (Description Below)
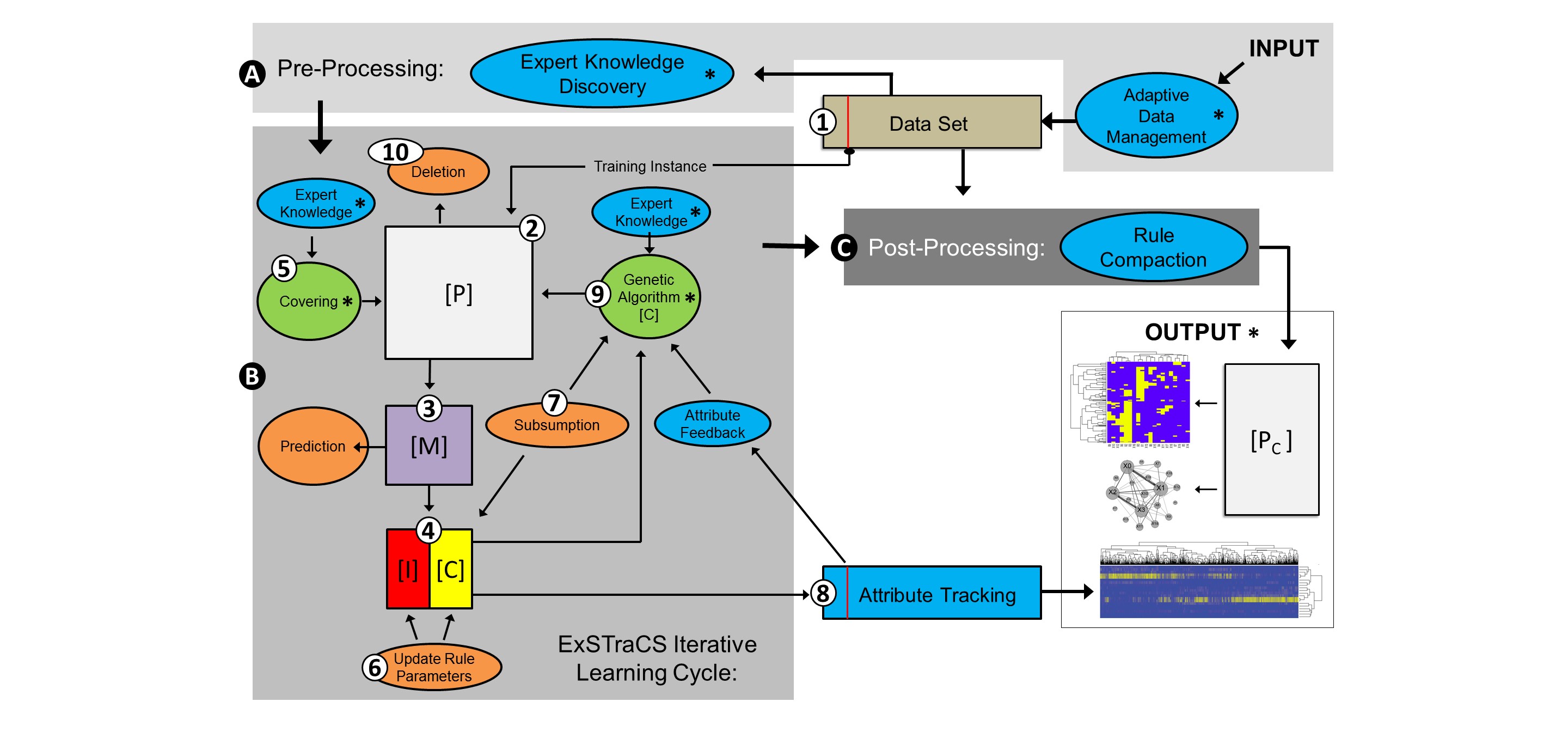
Following along with the figure given above, ExSTraCS begins with a data pre-processing stage (A). The algorithm takes as input, a configuration file and a finite training dataset that includes attributes and a single class endpoint variable. Adaptive data management determines and stores key characteristics of the dataset for later use. Next, expert knowledge (EK) attribute weights may be loaded or generated from the training data using one of the four included attribute weighting algorithms. Weights are utilized by the discovery mechanisms to favor the exploration of rules that specify attributes with higher weights. Attributes with higher weights should be more likely to be useful in discriminating between class/endpoints.
After pre-processing, the core iterative learning cycle (B) repeats through the following 10 steps until a maximum number of learning iterations is reached. (1) A single training instance is taken from the dataset without replacement. (2) The training instance is passed to a population [P] of classifiers that is initially empty. A classifier is a simple IF:THEN rule comprised of a condition (i.e. specified attribute states), and what is traditionally referred to as an action(i.e. the class phenotype). (3) A match set [M] is formed that includes any classifier in [P] that has a condition matching the training instance. (4) [M] is divided into a correct set [C] and an incorrect set [I] based on whether each classifier specified the correct or incorrect class phenotype. (5) If after steps 3 and 4, [C] is empty, covering applies expert knowledge to intelligently generate a matching and ‘correct’ classifier that is added to [M] and [C]. (6) For every classifier in [P], a number of parameters are maintained and updated throughout the learning process. Classifier parameters (e.g. fitness) are updated for classifiers within [C] and [I]. (7) Subsumption provides a direct generalization pressure that can merge overlapping rules in [C]. A similar subsumption mechanism is also applied to new classifiers generated by the genetic algorithm (GA). (8) Classifiers in [C] are used to update attribute tracking scores for the current training instance. These scores can be fed back into the GA to bias discovery toward useful attribute combinations. (9) The GA uses tournament selection to pick two parent classifiers from [C] based on fitness (i.e. a niche GA) and generates two offspring classifiers which are added to [P]. The GA includes two discovery operators; uniform crossover and mutation. (10) Lastly, whenever the size of [P] is greater than the specified maximum, a deletion mechanism selects a rule and either decrements the numerosity of the rule (if there are multiple copies), or removes it from [P] entirely if it is the only copy.
After all learning iterations have completed, rule compaction is applied as a post-processing step (C) to remove poor and/or redundant rules from [P] to yield a [Pc]. After completion, the ExSTraCS output may be used to make class predictions or evaluated and visulatized to facilitate knowledge discovery.